Abstract
Background: Early T-cell precursor acute lymphoblastic leukemia (ETP-ALL) is an aggressive hematological malignancy with poor prognosis after chemotherapy compared to other ALLs. ETP-ALL cases typically display a high burden of chromosomal copy number variations (CNVs) and myeloid gene mutations. To enhance the understanding of this disease, a computational biological modeling approach was developed to classify ETP-ALL vs. non-ETP-ALL patient based on genomic abnormalities and protein biomarkers associated with ETP-ALL and non ETP-ALL profiles.
Methods: Genomic profiling data for whole exome sequencing (WES), array CGH (Agilent) and karyotype data of 69 ETP- ALL and Non-ETP-ALL patient were used for this study. The genomic data for each patient were entered into a computational biology model (CBM) (Cellworks Group), which generates a disease-specific protein network map using PubMed and other online resources. CBM was used for a) Classification between ETP-ALL and Non-ETP ALL model, b) Identification of unique patient characteristics and c) Drug response.
For the CBM approach, we divided the data into training and validation data sets. 40 patients were selected for training data and 39 patients selected for the validation data set. In a training dataset, genomic data from 22 ETP-ALL patients were used from a GEO database (GSE28703) (N=11) and a prior genomics report (PMID:22237106) (N=11). The training dataset also included non-ETP-ALL patients from a GEO database (GSE28703) (N=18). Each patient's data was analyzed by CBM, to translates genomic variants into protein networks.
To validate these findings, genomic data from 15 ETP-ALL patients, different than training set patients, were used from a GEO database (GSE28703) (N=1), Moffitt Cancer Center (IRB201600284) (N=6), a prior genomics report (PMID: 22237106) (N=7), and an ETP-ALL LOUCY cell line with known genomic aberrations. The validation dataset also included Non-ETP-ALL patients different than training set patients, from a GEO database (GSE28703) (N=14).
Results: Prediction accuracy for classification between ETP-ALL and Non ETP ALL patient was 89.65%. PPV value was 86.67% and NPV value was observed 92.86%. Sensitivity and Specificity of prediction outcome was observed as 92.86% and 86.67% respectively.
The simulation model identified six protein biomarkers which predicted increased activity in ETP-ALL profiles compared to non-ETP-ALL cases: STAT3, RUNX1, HIF1A, FOXM1, CEBPA, mTORC1.
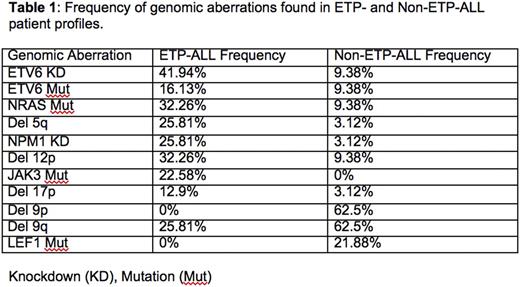
Key genomic differences were also identified between ETP-ALL and non ETP-ALL patients (Table1).
Conclusions: ETP-ALL is distinct from other T-ALL with respect to genomic and protein network abnormalities. Computational biology model can accurately classify between ETP-ALL and Non ETP-ALL patients. CBM also predicted unique biomarkers between ETP-ALL and Non-ETP-ALL patients.
Kumar: Cellworks: Employment. K: Cellworks: Employment. Vasista: Cellworks Research India: Employment. Tyagi: Cellworks: Employment. Rajagopalan: Cellworks: Employment. Alam: Cellworks: Employment. Abbasi: Cellworks Group Inc.: Employment. Vali: Cellworks Group Inc.: Employment. Cogle: Celgene: Other: Membership on Steering Committee for Connect MDS/AML Registry.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal